Molecules and Meaning:
How Do Molecules Become Biochemical Signals?
Bernard Testa1,
Lemont B. Kier2 and Andrzej J.
Bojarski3
1) Institute of Medicinal Chemistry, School of Pharmacy,
University of Lausanne, CH-1015 Lausanne, Switzerland, <Bernard.Testa @ ICT.UNIL.CH>
2) Center for the Study of Biological Complexity, Virginia
Commonwealth University¸ Richmond, VA 23298, USA, <kier@hsc.vcu.edu>
3) Department of Medicinal Chemistry, Institute of
Pharmacology of the Polish Academy of Sciences, 12 Smetna St., PL-31343 Krakow,
Poland,
<bojarski@rabbit.if-pan.krakow.pl>
©This paper is not for reproduction, even partial, without
the express permission of the first author.
ABSTRACT
The objective of this paper is to reflect on how molecules
can acquire macroscopic meaning (i.e., carry a message to macroscopic levels)
in a context of biological evolution. First, the structure of molecules is
explained in terms of form (molecular geometry), function (measurable or computable
molecular properties), and fluctuation. Fluctuations in form and function
create distinct molecular states, and the ensemble of all molecular states
defines a molecular space (also known as a property space).
The second part examines molecules in a chemical context. The
interplay between a chemical compound and its environment creates a complex
system in its own right, as exemplified by solutions. A solute influences the
solvent by affecting its organization and some colligative properties, while
the solvent often has a marked influence on the solute by constraining its
property space and so selecting some of its molecular states. Solutions may
display emergent properties not existing in the separate components, e.g.
chemical reactivity, implying that information has been created upon formation
of the complex system.
The third part of the paper discusses the interaction of
chemical compounds with biological media. In contrast to abiotic environments
such as solvents whose degree of organization is comparatively low, biological
media are characterized by a high degree of organization. Examples at the
macromolecular level include functional proteins (receptors, enzymes,
transporters, ...) or nucleic acids. When a molecule is recognized by such a
macromolecule and interacts (binds) productively with it, a complex system is
produced whose emergent property is the functional response, and which strongly
constrains both of its components. The chemical is frozen into a single or a
very limited number of molecular states (induced fit), whereas the
macromolecule is activated by a conformational change (e.g. an allosteric
effect). Here again, emergent information appears in the complex. However,
there is an essential difference with abiotic systems since the emergent information
can now be translated into a functional biochemical reaction that in turn will
be amplified into a macroscopic biological response. In other words,
information emerging in the molecule-macromolecule complex is a signal that
becomes meaning as it is recognized in the higher hierarchy of nested
biological contexts.
1
Preamble: signal molecules
Chemical signals were the first means of long-distance
communication evolved by living organisms, and they remain among the most
effective and specific ones. Thus, pheromones are a major means of communication
between conspecific organisms, whereas different species can interact via
chemical signals that elicit attraction, repulsion, cooperation, etc. Chemical
regulation within multicellular organisms is particularly well documented and
involves hormones, neurotransmitters and other regulators. Drugs are another
case in point, being messages sent to ailing cells and organs and aimed at
correcting pathological states (Testa 1996, 1997). A list of signal molecules
therefore includes:
·
poisons and
repellents (inter-species)
·
attractors
(inter- and intra-species)
·
pheromones
(intra-species)
·
neurotransmitters,
hormones, growth factors (intra-organisms)
·
drugs
The overall molecular mechanism by which information
is transmitted from organism to organism or from cell to cell is always the
same, namely:
·
Emitted signal
molecules are recognized by and fit into specific receptors (The key enters
the lock).
·
The formation of
the receptor-ligand complex alters the state of the receptor (The lock is
turned).
·
This triggers a
biochemical cascade (The opening mechanism is activated).
·
The ultimate
outcome is a macroscopic response (The door opens).
The problem we examine here is how molecules can
acquire specific messenger functions. Stated differently, how could Evolution
endow molecules with biological meaning? The evolutionary explanation we
propose is based on the facts that a) molecules exist within their property
space; b) the molecular environment (e.g. a receptor) interacts with the molecules
by constraining their property space (dissolvence) and so selecting some of
their molecular states; c) in turn, the molecules act on the receptor by
modifying it; d) this mutual adaptation creates a molecule-receptor complex,
i.e., a complex system whose functional response is an emergent property; and
e) this functional response is a signal which becomes meaning in the
higher levels of complexity — the biological context.
2
molecular structure and property space
2.1
The concept of molecular structure
The description of molecules may be approached by
considering form, function, and fluctuation (Testa and Kier 1991; Testa et al.,
1997). Molecular form ("what a molecule is") can be equated
with molecular geometry, namely atom connectivity (2D-structure) and more realistically
the 3D-structure. Components of molecular form are called structural
attributes. Molecular function ("what a molecule does") is
interpretable from observations made during experiments and is expressed as
measurable or computable properties. Structure and properties (i.e., form and
function) influence each other and are indubitably intertwined. The third component
in this approach is molecular dynamics, namely the fluctuation in form
and function (Prigogine, 1978).
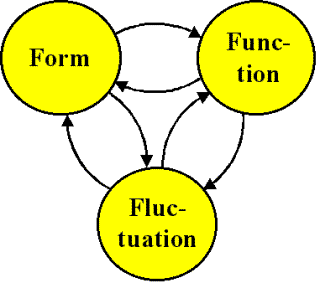
Figure 1: A comprehensive representation of molecular
structure in the broadest sense, viewing form, function and fluctuation as its
three essential components. A chemical compound can exist in a number of
molecular states differing in conformation, surface area and volume, H-bonding
capacity, polarity, lipophilicity, etc.
Form, function and fluctuation cannot be ordered
causally or hierarchically. Rather, they are viewed as being of equal
importance and feeding on each other, as schematized in Figure 1. Fluctuation
influences form. To give an example, consider how in some compounds a labile
hydrogen can jump from one position to another. This changes the atom
connectivity (2D-geometry) of the molecule, which experiences a prototropic
equilibrium and thus fluctuates between two or more states known as tautomers.
Similarly, the 3D-geometry of a molecule can vary markedly depending on its
flexibility, resulting in stereoisomers separated by low-energy barriers and
well known to chemists as conformers (conformational isomers). While
tautomerism is restricted to a relatively limited number of compounds and
involves two (seldom three) tautomeric states, conformational isomerism is a
phenomenon of very frequent occurrence that produces a great many (an infinity
depending on definition) conformational states. The ensemble of these states
defines the conformational space, also known as the conformational hypersurface
of a compound.
Form influences fluctuation. This is a rather
trivial statement considering that the capacity of a molecule to oscillate between,
e.g., tautomeric or conformational states is entirely pre-determined by its
chemical constitution. This makes it clear that form and fluctuation are interdependent
and influence each other, in complete similarity with the interdependence between
form and function.
Function and fluctuation also influence each other.
This is a conclusion that derives logically from the above statements, and
which can easily be seen in chemical examples. That tautomers display different
chemical properties is again well known to chemists. Similarly, it is common
chemical knowledge that electronic properties (e.g. ionization state) will
influence conformational behavior.
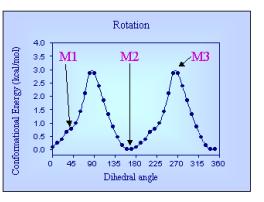
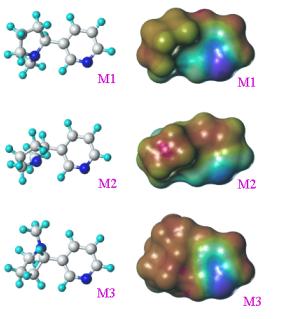
Nicotine is taken as an example here to illustrate
the above principles. This compound has but one rotatable bond, and its
conformational behavior is mainly described by the relative free energy associated
with each value of the dihedral angle (Figure 2). Three individual conformers
are shown in Figure 2, M2 being the most populated one since it corresponds to
the global energy minimum, M3 corresponding to the rotation barrier, and M1
being of intermediate energy and probability. As mentioned above, the three
conformers have clearly different forms (just consider the relative location of
the two nitrogen atoms, but they also have different properties as exemplified
by their different electrostatic potentials.
The general conclusion at this stage is therefore
that form, function and fluctuation are interdependent. They depend on each
other in a quantitative manner, and in some cases even qualitatively. This
systemic and global perception of molecules has implications that will become
evident below.
2.2
Molecular states and property space
Molecular fluctuation delineates the ensemble of all
probabilistic changes a molecule can undergo in form and function. This
generates a very large number of molecular states, which are snapshots
of the molecule at a given moment in time. Each state is characterized by a
unique combination of geometry (form) and associated properties (function).
Reciprocally, any property exhibited by a compound will thus have a distinct
value for each molecular state occupied by that compound.
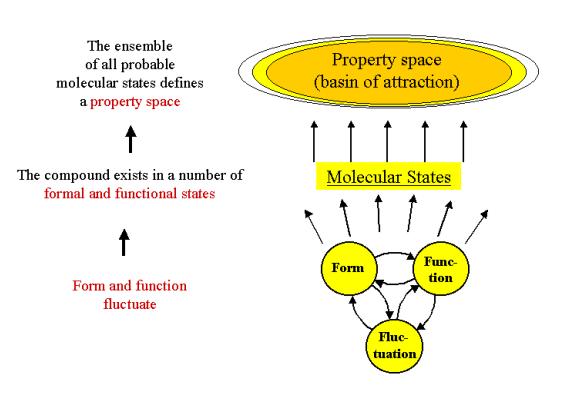
Figure 3: Molecular states are the expression of the
mutual interdependence of form, function and fluctuation. The ensemble of all
molecular states of a compound defines its property space, namely the range of
values each property can span.
The ensemble of all possible states will span a
range of values for all properties, thus delineating a property space.
The latter can also be conceived as the basin of attraction of the property
states of a compound. The concept of a basin of attraction can be schematized
in pictorial language as shown in Figure 3. A physically more realistic
representation of a basin of attraction of all molecular states is afforded by
an energy landscape, namely a hypersurface whose dimensions are the
energy of the system, plus all its other variables (Testa et al 1997). Usually,
and this is the convention also adopted here, the more probable states of a
molecule (i.e., its states of lowest energy) are represented as valleys in the
energy landscape, whereas the states of highest energy are represented by peaks
and the transition states as mountain passes. There is an energy maximum beyond
which the molecule breaks down and ceases to exist, explaining why an energy
landscape is finite. A schematic representation of an energy landscape will be
shown later (see Figure 5 below). Note that any complex system could a priori be represented by an energy
landscape, but the hyperdimensionality increases incommensurably for systems of
higher complexity.
A molecular property of great biological relevance
is lipophilicity, namely the preferential affinity of a solute for
lipid-like over water-like solvents. This property is commonly measured, but it
can also be reliably computed from 2D– and 3D–structures (Carrupt et al 1997).
In particular, an algorithm known as the Molecular Lipophilicity Potential
(MLP) calculates a virtual lipophilicity for each conformer. The results have
revealed large differences between the various conformational states of a
compound, up to one order of magnitude or even more (Testa et al 1996). This
phenomenon, which is particularly marked for large molecules such as various
drugs and biomolecules containing both hydrophilic and hydrophobic groups (Jiang
1998), has been termed the chameleonic effect (Carrupt et al 1991).
It follows from the above that one way to rank all
molecular states of a compound is along an axis of polarity, as will be done
below in Figure 5.
3
Molecules in a chemical context: External constraints
3.1
The molecule-medium combination as a complex system
We now examine the interplay between a molecule and its
molecular environment, showing that it creates a complex system in its own
right. We note that a complex system results from interactions (also
called transactions) between its components, and that it exhibits emergent
properties non-existent in its components (Capra 1983). A third (and
hitherto not explicited) feature of complex systems, that of dissolvence, will
be discussed later.
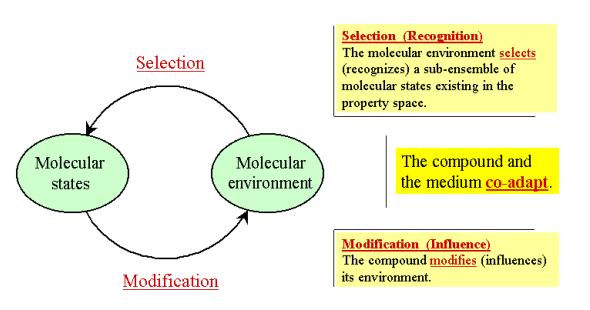
Figure 4: The complex system formed by a chemical
compound and its environment results from two types of interactions between its
components. The molecular environment selects a sub-ensemble of molecular
states in the compound, whereas the latter modifies its environment.
As schematized in Figure 4, the molecule and its
environment influence each other. At the macroscopic level, it can be stated
that a compound modifies the medium with which it is in contact, as seen for
example with changes in physical properties of a solution relative to the pure
solvent, a modified fluidity in a membrane, or an allosteric effect in a
protein.
But the medium also influences the compound. For
example, a solvent will influence the electronic properties of the solute,
which may exhibit changes in its color and UV spectrum. Similarly, the
conformational behavior of the solute is markedly affected by the medium.
In the perspective of Figure 4, the molecule and its
environment co-adapt to each other within their property space, a
phenomenon that can also be viewed as a reversible co-evolution. In this
writing, we focus essentially on the influence of the medium on the compound it
engulfs. As shown below, this influence involves selection by the environment
of a fraction of the property space accessible to the molecule. Such changes in
property space have been termed dissolvence, being considered as the
counterpart of emergence (Testa and Kier 2000).
3.2
Solvent constraints
on the property space of solutes
Figure 4 not only schematizes the transactions between a
molecule and its environment, it also raises the question of the intensity of
their mutual adaptation. A precise answer appears impossible, but the wealth of
available experimental evidence ascertains a qualitative trend. Indeed, the
degree of mutual adaptation between a compound and its environment depends
mostly on the degree of organization of the latter. Here, we examine the case of
a solvent, i.e., a medium with a low degree of organization. Biological media,
which are characterized by a relatively high (membranes) or even an extremely
highly (functional proteins) degree of organization will be discussed in
Section 4.
Solvents have a rather high degree of macroscopic
(apparent) order, but at the molecular level large random movements and
fluctuations take place. The degree of mutual adaptation between solute and solvent
will be comparatively low. The solute will have some influence on the solvent,
e.g. by local alterations of its structure (e.g. the hydrophobic effect), and
by altering slightly some colligative properties such as its freezing point,
boiling point, vapor pressure and viscosity. As for the solvent, it usually has
a marked influence on the properties of the solute. What is clearly revealed by
experimental and computational investigations, for example, is the effect of
the solvent on the conformational behavior of the solute, resulting in the selection
of some among all the possible molecular states (Testa and Bojarski 2000; Testa
et al 1999).
As a general rule, polar (hydrophilic) solvents will
favor the more polar conformations and states, whereas the less polar
(lipophilic) solvent will favor less polar conformations and states. In short,
"Like selects like", a statement that extends the meaning of one of
the most basic rules in chemistry, "Similia
similibus solvuntur" (Like dissolves like).
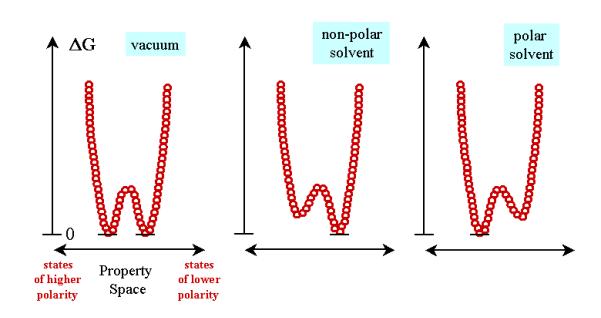
Figure 5: This figure shows the property space of a hypothetical
compound as influenced by the mo-lecular environment. Each recognizable state
of the molecule is represented by a small cir-cle whose position is defined by
its relative energy (ordinate axis) and its relative polarity. In this
representation, the property space is condensed to an axis of polarity, as
explained in subsection 2.2. The compound is taken to have two low-energy
states which are of equal energy/probability in the vacuum. In a solvent,
however, "like selects like".
The experimental rule "Like selects like"
can be depicted by Figure 5, which shows the property space of a hypothetical
compound. Each recognizable molecular state is represented by a small circle
whose position is defined by its relative energy (ordinate axis) and its relative
polarity. In other words, the property space is reduced to and represented by
an axis of polarity, as explained in subsection 2.2. The compound is seen to
have two low-energy states of equal energy/probability in the vacuum. In a
non-polar solvent, the solute will exist predominantly as conformers of lower
polarity, whereas the opposite is true in polar solvents.
3.3
Mutual adaptation and the emergence of information
As ambiguously encapsulated by Gel-Mann (1995),
"Information is concerned with the selection from alternatives". This
statement is also valid for the phenomenon described above, namely the
recognition and selection by a solvent of some among all existing molecular
states of a solute. But where does the necessary information originate from?
Exclusively from the solvent, in which case all solutes would be identically
constrained in a given solvent? Or exclusively from the solute, in which case
the constraints experienced by a given solute would be solvent-independent?
Taken alone, none of these two explanations is
satisfactory. In contrast, the information present in the separate components
is mutually perceived when the chemical compound and the solvent encounter each
other, interact and form the solute-solvent complex (the solution). This
results in the emergence of new properties, in other words in the emergence of
information.
The example of chemical reactivity can also serve to
illustrate this phenomenon. Chemical reagents are seldom or poorly reactive as
isolated molecules in the vacuum, but they need an adequate milieu to express
their reactivity (i.e. to be "activated"). In fact, we are dealing
here with a three-component system, since the reagent and its chemical target
need an adequate milieu to enter the chemical reaction. There is also another
deduction to be made from this example, that the property space of reagents is
constrained toward increased reactivity. In this sense, it would be misleading
to equate dissolvence with mere deprivation; rather, dissolvence implies
constraints on (changes in) property space that cannot a priori be qualified as negative or positive. This however will
not be discussed in the present writing. Suffice it to repeat that the
information necessary for a chemical reaction to proceed readily emerges in the
reagent-milieu complex from information present in the components and upon
their co-adaptation, as depicted schematically in Figure 6.
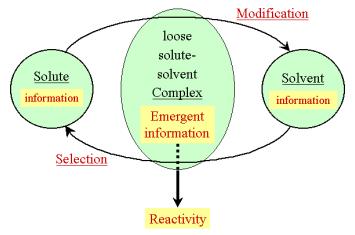
Figure 6: Scheme depicting the emergence of information
upon formation of a solute-solvent complex system.
4
molecules in a biological context: the emergence of meaning
4.1
molecular machines
In section 3, we have seen how a non-biological (abiotic)
medium with a relatively low degree of organization constrains a solute. We now
turn our attention to biological media with an intermediate or high degree of
organization. These are the media encountered by signal molecules such as
hormones, neurotransmitters and drugs before and when they reach their sites of
action.
Media with an intermediate degree of organization
are for example biological membranes through which bioactive compounds must
permeate to reach their targets. Here, we postulate an intermediate selection
of molecular states and an intermediate degree of influence on the medium.
Media with a high degree of organization are of great biological and
pharmacological significance, being functional macromolecules such as
receptors, nucleic acids, enzymes and transporters. The remainder of this text
is restricted to the interaction of biomolecules with such highly organized
biological media.
Functional biomacromolecules have a broad property
space characteristic of their chemical integrity (i.e., their primary structure
= the sequence of their monomers) which does not concern us here. But they have
a second, very narrow property space with a rigidly defined tertiary structure
(i.e., their 3D-structure) which contains the single or the very few macromolecular
state(s) compatible with functionality. Structural water (i.e., specifically
bound water molecules) plays an essential role in stabilizing such specific
states, not to mention a functional role in some enzymatic reactions (Kier and
Testa 1996). Thus, fluctuations within the narrow property space of a
functional macromolecule are compatible with its function, whereas fluctuations
outside this narrow property space but inside the broader space destroy
functionality while preserving chemical integrity.
The reason for such a remarkably high constraint on
their 3D-structure is that macromolecules do not function in isolation but as
constituents of functional assemblies some scientists like to call "molecular
machines". These are complex systems of higher order resulting from
transactions between functional macromolecules which recognize each other based
on the stereoelectronic properties of exquisitely fine-tuned complementarity
sites.
4.2
The ligand-receptor complex and its functional response
For the sake of simplicity, the following discussion will
be restricted to receptors, bearing in mind — mutatis mutandis — that it also concerns other molecular machines.
Besides specific sites for macromolecule-macromolecule
recognition, other high-affinity sites exist on receptors and other functional
macromolecules to recognize and bind bioactive compounds. Here however, and in
contrast to the loose solute-solvent interactions discussed above, strong
transactions occur between the ligand (i.e., the bioactive compound), the macromolecule
and structural water molecules. Such transactions are the mechanism by which a
ligand-macromolecule complex is formed.
On the one hand, the ligand markedly influences the
macromolecule by activating it (e.g. by an allosteric effect; Kenakin 1996) and
triggering its biological response (in the case of an agonists or substrate),
or by blocking its functioning (in the case of an antagonist or inhibitor). On
the otehr hand, the influence of the receptor on the ligand is a remarkably
strong one since a very narrow range of molecular states are selected, usually
a single conformation. This is the well-known phenomenon of the induced fit
(Koshland 1976). As long as the ligand-receptor complex lasts, the ligand
remains practically frozen in a single conformational state, a strong
constraint that allows for a tight complex and, we postulate, for an effective
and selective biochemical response. Here like in solute-solvent systems, the
information present in the separate components is mutually perceived as the
components recognize each other, co-adapt, and form the ligand-receptor system
(Figure 7).
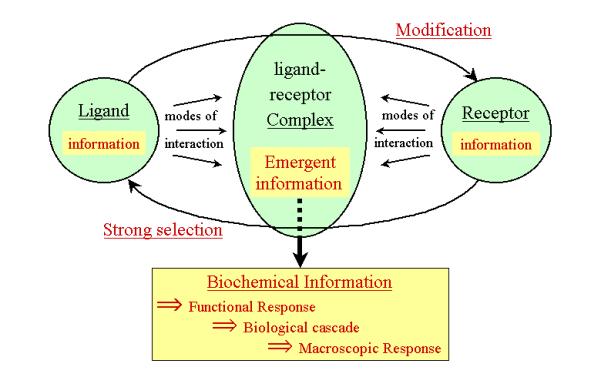
Figure 7: Scheme depicting the emergence of information
upon formation of a functional ligand-receptor complex system. The activated receptor
generates a biochemical response that will be amplified downstream into a
macroscopic biological response.
In this co-adaptation, the receptor selects a
restricted number of molecular states in the ligand, whereas the latter influences
(modifies) the latter. Feedback control also exists in such a system, which
thus becomes a model of the flow of information in a functional ligand-receptor
complex. In such a simplified model, neither the ligand nor the receptor alone
can produce a complete functional response, the functional response being
indeed an emergent property of the ligand-receptor complex.
4.3
Nested biological contexts and the emergence of meaning
As discussed in the Preamble of this writing, a variety of
bioactive compounds have a signal function. There is here an essential
difference with abiotic systems since the emergent information can be translated
into a functional biochemical reaction that in turn will cascade and be
amplified into a macroscopic biological response. Figure 7 depicts the
emergence of biochemical information, but it falls short of explaining how
bioactive molecules can trigger macroscopic responses, in other words how they
become endowed with macroscopic biological meaning.
In an enlightening text, Cohen and Stewart (1995)
have explained and exemplified that no message exists that would contain
inherent information independent of the context. One of their examples is
particularly inspiring:
"When you transmit "Make me a pearl"
to an oyster, the message takes the physical form of a tiny piece of grit but
produces a wonderful, lustrous result. [.......] Meaning is a quality, not a quantity,
and is highly dependent upon context."
How is this relevant to our discussion? When the
piece of grit enters the oyster, it triggers a biochemical reaction in some
cells and forces them to produce a solid secretion known as mother-of-pearl.
However, the entire biological context, namely the oyster, is needed to produce
a pearl from the mother-of-pearl, in other words to "inform" the
mother-of-pearl. As made clear by Cohen and Stewart, if all the information
needed to make a pearl were encoded in the tiny piece of grit, the latter
should contain an amazing amount of it. In fact, the piece of grit only
contains the information to interact with cells. The latter have the latent
capacity to secrete mother-of-pearl, but they need a stimulus. Mutual recognition
and interaction between grit and cells must occur for a functional response to
be produced, in this example for mother-of-pearl to be secreted. However, only
the oyster can give form to the mother-of-pearl and transform it into a pearl.
Stated differently, the oyster is the biological context that gives meaning to
the mother-of-pearl by creating a pearl.
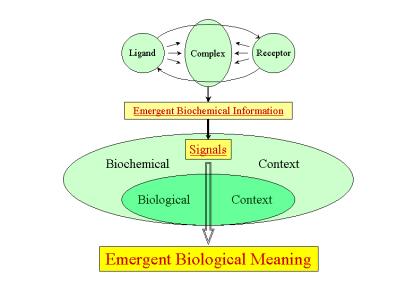
Figure 8: Scheme depicting the emergence of meaning
resulting from the interaction between bio-chemical information (the functional
response of the ligand-receptor complex) and a hier-archy of nested biological
contexts.
Returning to bioactive compounds and their
receptors, we can expand Figure 7 by stating that while biochemical information
emerges in the ligand-receptor complex, it needs higher levels of biological
complexity to become macroscopically meaningful. These levels of higher
complexity are the biological context, or better said a hierarchy of nested
biological contexts (organelles, cells, tissues, organs, systems, organisms,
...). At each level, recognition and transactions must occur for the
information to become a meaningful signal recognized by the next contextual
level, and so on. The resulting spiral is depicted in Figure 9 and leads to the
deduction that the autoregulation of organisms is based on a genuine
bootstrapping of information and meaning.
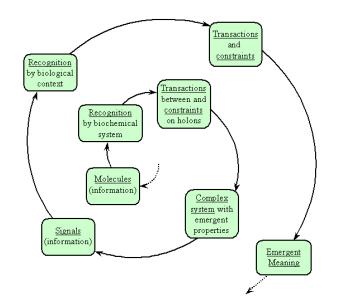
Figure 9: The bootstrapping of biological information
and meaning, as deduced from the actualization of information occurring in
nested biological contexts.
5
conclusion: emergence and dissolvence in the evolution and
autoregulation of living systems
To summarize, we have seen that constraints on the
property space of molecules and macromolecules (dissolvence) are inherent in
biochemical recognition, which initiates the transactions between components
(holons) that allow a complex system to be created (Figures 1–5).
The existence of such constraints reveals an
information flow between the holons and the complex system. The information
present in the holons allows their mutual recognition and the creation of the
complex system. (Figure 6).
In the complex system, information emerges as a
response, often in the form of a signal sent outward. Such signals will be
recognized by and interact with the next level of biological complexity
(Figures 7–8).
The spiral of dissolvence and emergence thus
progresses in a hierarchy of nested biological contexts (Figure 9). Levels
after levels of biological meaning are thus created by bootstrapping,
propelling the evolution of living systems toward levels of ever increasing complexity
(Figure 10).
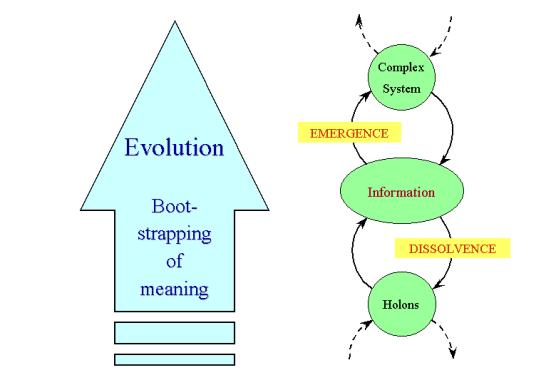
Figure 10: The coupled processes of dissolvence and
emergence are the channels by which information is exchanged between holons and
complex systems. Biological meaning emerges by boot-strapping, which propels
the evolution of living systems toward ever increasing levels of complexity.
References
Capra, F. 1983. The Turning Point. Science, Society and
the Rising Culture, p. 285-332. London: Fontana Paperbacks.
Carrupt, P.A., Testa, B., Bechalany, A., El Tayar, N.,
Descas, P. and Perrissoud, D. 1991. Morphine-6-glucuronide and
morphine-3-glucuronide as molecular chameleons with unexpected lipophilicity.
Journal of Medicinal Chemistry 34:
1272-1275.
Carrupt, P.A., Testa, B. and Gaillard, P. 1997.
Computational approaches to lipophilicity: Methods and applications. in K.B.
Lipkowitz and D.B. Boyd, eds. Reviews in Computational Chemistry, Vol. 11, pp.
241-315. New York: Wiley-VCH.
Cohen, J. and Stewart, I. 1995. The Collapse of Chaos -
Discovering Simplicity in a Complex World, pp. 288-293. New York: Penguin
Books.
Gell-Mann, M. 1995. The Quark and the Jaguar, p. 37.
London: Little, Brown & Co.
Jiang,
X.-K. 1988. Hydrophobic-lipophilic interactions. Aggregation and
self-coiling of organic molecules.
Accounts in Chemical Research 21:
362-367.
Kenakin, T. 1996. Receptor conformational induction
versus selection: all part of the same energy landscape. Trends in
Pharmacological Sciences 17:
190-191.
Kier, L.B. and Testa, B. 1996. Complex systems in drug
research. II. The ligand-active site-water confluence as a complex system.
Complexity 1(4): 37-42.
Koshland Jr., D.E. 1976. Role of flexibility in the
specificity, control and evolution of enzymes. FEBS Leters 62: E47-E52.
Prigogine, I. 1978. Zeit, Struktur und Fluktuationen
(Nobel Lecture). Angewandte Chemie 90:
704-715.
Root-Bernstein, R.S. and Dillon, P.F. 1997. Molecular complementarity I:
the complementarity theory of the origin and evolution of life. Journal
of Theoretical Biology 188: 447-479.
Testa, B. 1996. Missions and finality of drug research:
A personal view. Pharmaceutical News 3:
10-12.
Testa, B. 1997. Drugs as chemical messages: Molecular
structure, biological context, and structure-activity relationships. Medicinal
Chemistry Research 7: 340-365.
Testa, B. and Bojarski, A. 2000. Molecules as complex
adaptative systems. Constrained molecular properties and their biochemical
significance. European Journal of Pharmaceutical Sciences 11: S3-S14.
Testa, B. and Kier, L.B. 1991. The
concept of molecular structure in structure-activity relationship studies and
drug design.
Medicinal Research Reviews 11: 35-48.
Testa, B. and Kier, L.B. 2000. Emergence and dissolvence
in the self-organization of complex systems. Entropy 2: 1-25. http://natsci.net/reprints/entropy/2000/e2010001.pdf or
http://www.mdpi.org/entropy/papers/e2010001.pdf
Testa, B., Carrupt, P.A., Gaillard, P., Billois, F. and
Weber, P. 1996. Lipophilicity in molecular modeling. Pharmaceutical Research 13: 335-343.
Testa, B., Kier, L.B. and Carrupt, P.A. 1997. A systems
approach to molecular structure, intermolecular recognition, and
emergence-dissolvence in medicinal research. Medicinal Research Reviews 17, 303-326.
Testa, B., Raynaud, I. and Kier, L.B. 1999. What
differenciates free amino acids and amino acyl residues ? An exploration of
conformational and lipophilicity spaces. Helvetica Chimica Acta 82: 657-665.